-
Genomic DNA damage can be induced by different physical or chemical endogenous and exogenous factors. Cells respond to DNA breaks for survival[1]. Those proliferating cells have to arrest the cell division cycle to repair DNA lesions. If the repair cannot be completed, cells undergo programmed cell death. Ionizing radiation(IR) and chemical agents can cause a variety of DNA lesions, namely, mutation, single-strand, and double-strand breaks. Double-strand breaks (DSBs) are the most serious damage type of genomic DNA and individual DSBs for cells are fatal. Cells use two major repair pathways, namely, homologous recombination and non-homologous end joining(NHEJ), to complete DSB repair. Homologous recombination requires an undamaged homologous DNA template to replace the adjacent broken high-fidelity DNA[2]. By contrast, NHEJ is an error-prone quick-repair process that directly connects the ends of two broken strands[3].
The sequencing of the human genome showed that only approximately 20 000 protein-coding genes exist, representing < 2% of the total genomic sequence, whereas the rest of the genome has over 90% transcribed into non-coding RNAs(ncRNAs). Initially, these ncRNAs were called "transcriptional noise". Then, the development of modern molecular biology techniques enabled scientists to discover that ncRNAs were involved in many biological processes, such as chromatin remodeling, histone modification, DNA methylation, amplification, and rearrangement, among others. The regulation pattern of RNA molecules is highly diverse. NcRNAs can be roughly classified into the following two categories based on the length of ncRNAs: Long non-coding RNAs(LncRNAs) greater than 200 bp, and short non-coding RNAs less than 200 bp. The latter includes micro RNA, PIWI(P-element induced wimpy testis)-interacting RNA, endogenous small interfering RNA, and competitive endogenous RNA. This diversity creates a highly enriching, rigorous, and orderly gene expression process.
In recent years, studies have shown the participation of ncRNAs in nearly all biological processes and signaling pathways. DNA damage and repair in the DNA damage response(DDR) process further require the ncRNAs regulation. Some evidence show that DSB repair proteins, including 53BP1(p53 binding protein 1), BRCA1(breast cancer 1), and Ku70(a protein that, in humans, is encoded by the X-ray repair cross-complementing 6 gene), combine ncRNAs into DSB sites, thereby affecting the efficiency of repair[4]. Micro RNA expression is regulated by DNA damage, and is further directly affected by DDR proteins in certain cases[2-3, 5-6]. However, how these ncRNAs affect DDR remains unclear.
LncRNAs are transcribed RNA molecules with a length of over 200 nucleotides. At present, scientific research mainly focuses on a role of lncRNAs in cancer and other major diseases, but the regulatory mechanism in the development of DDR remains unclear. Wang et al.[7] found that a group of lncRNAs, CCND1(cyclin D1), regulate DDR caused by IR through epigenetic modification of chromatin. Since then, other researchers have reported lincRNA-p21(also called TP53COR1, tumor protein p53 pathway corepressor 1), PANDA(promoter of CDKN1A antisense DNA damage activated RNA), and DDR-related lncRNAs. LncRNAs may control DDR through other mechanisms. Still, no systematic study has been conducted on this aspect. The interplay among lncRNAs, microRNAs, and proteins in the regulation of DNA damage response requires investigation. In radiobiology, research on IR-induced DDR regulated by lncRNAs remains extremely rare. Thus, this paper reviews the emerging role of lncRNAs in DNA damage repair.
-
Genomic instability is one of the most common features of tumor cells, which may be caused by the following: DNA injury, comprehensive effect of tumor-specific DNA repair defects, and failure of cell cycle arrest before the cell divides into daughter cells. To maintain genomic stability and respond to DNA damage, cells evolve a set of complex cellular responses to DDR, thereby coordinating DNA damage checkpoints of the cell cycle[1].
The matter of coordinating DDR is divided into sensor, transducer, and effector. DNA damage recognition is the first step in activating DNA-damage checkpoint signaling cascades. DNA damage is detected by corresponding sensor proteins, such as MRN(MRE11-RAD50-NBS1)complexes that receive signals; the RPA (replication protein A) protein binds to a single-strand DNA at the DNA damage loci. Subsequently, the highly conserved DDR kinase, ATM(ataxia telangiectasia-mutated), and ATR(ataxia telangiectasia and Rad3 related), have been recruited and activated. In yeast and mammalian cells, Tel1/ATM identifies DSBs, whereas Mec1/ATR can be activated when a long single strand of DNA ends exists. Once activated, the DDR signal is transducted via the phosphorylation and activation of downstream kinase, such as CHK1(checkpoint kinase 1) and CHK2(checkpoint kinase 2). Checkpoint activation triggers a series of cellular responses, allowing cells enough time to repair the DNA damage by coordinating cell-cycle progression.
-
LncRNAs may have the following four functions in the DDR pathway: (1)guiding or signaling recruiting repair proteins or chromatin-modifying complexes to sites of DNA damage, (2)acting as scaffolds for DNA repair proteins or chromatin remodeling machinery at the site of the DNA repair foci, (3)preventing the action of negative regulators of DNA repair at the site of DNA damage by acting as decoys, and (4)regulating DNA damage-sensitive gene expression programs, such as lincRNA-p21 and PANDA[8].
-
DDR is a major protective mechanism against genomic instability in eukaryotic cells. In the past couple of decades, several protein components have been identified in the DDR pathways, including DNA damage sensors, signaling PIKKs(phosphatidylinositol 3-kinase-related kinases), and effectors[1]. The ATM kinase is a primary PIKK that responds to DSBs and activates a variety of cell activities[9]. The ATM-p53 signaling plays a central role in cell-cycle checkpoints and cell death pathways following DNA damage and oncogenic stresses[10]. Regulators in the ATM-p53 signaling modulate not only the activity and stability of signaling proteins but also their mutual recognition, interaction, and signaling complex formation.
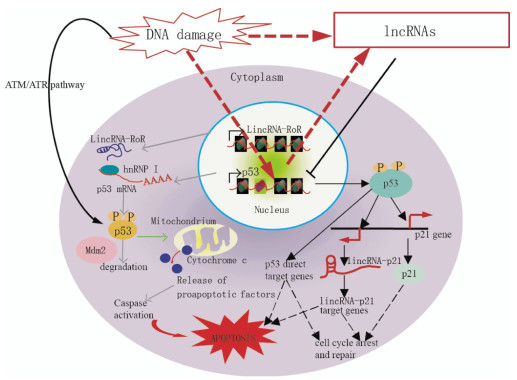
Aberrant expression of individual lncRNAs has been reported in tumors of various tissue origins[11]. Furthermore, recent data revealed that lncRNAs transcripts can modulate gene activity in response to DNA damage[12]. LncRNAs are increasingly associated with DDR. Fig. 1 shows the proposed mechanisms of lncRNAs-mediated regulation of the p53 pathway. Kitagawa et al.[13] emphasized that non-canonical DDR are participated by certain lncRNAs in cell cycle arrest or induct apoptosis, whereas the ATM/ATR pathway refers to a canonical DDR to deactivate CDK(cyclin-dependent kinase) activity as a DNA damage checkpoint. LncRNAs-mediated non-canonical pathways may ensure DDR, which is diverse and reliable depending on the cellular context.
Figure 1. Model demonstrating the proposed mechanisms of lncRNA-mediated regulation of the p53 pathway, which controls cell cycle arrest, repair, and apoptosis in response to DNA damage. In response to DNA damage, p53 is stabilized and activated via phosphorylation mediated by the ATM/ATR pathway. p53 directly binds the target genes and regulates their expression to control cell cycle arrest, repair, and apoptosis. p21 and lincRNA-p21, which are transcribed near the p21 gene, are p53-target genes. LincRNA-p21 controls the expression of some p53-target genes. p53 function is partially mediated by gene regulation via lincRNA-p21.
-
A number of lncRNAs are expressed in a cell cycle-regulated manner. Some of these lncRNAs are induced by DNA damage and inhibit cell cycle progression by regulating cell cycle regulators, such as ncRNACCND1, which suppresses Cyclin D1 transcription(G1 phase); gadd7(growth arrested DNA-damage inducible gene 7), which destabilizes CDK6(cyclin-dependent kinase 6) mRNA(G1 phase); ANRIL(antisense non-coding RNA in the INK4 locus), which suppresses p15/p16 transcription with PRC1/2(G1 phase); and PANDA, which suppresses FAS(Fas cell surface death receptor) and BIK(Bcl-2-interacting killer) transcription.
NcRNACCND1 or pncRNA(promoter-associated non-coding RNA) is transcribed from the upstream region of the cyclin D1 gene, CCND1, and negatively regulates cyclin D1. NcRNACCND1 functions as a transcription factor regulator[7]. It is induced in a DNA-damage dependent manner, and associates with and recruits TLS(translocated in liposarcoma)[14], an RNA binding protein. The ncRNACCND1-TLS complex is recruited to the CCND1 promoter to inhibit the activity of the coactivator, CBP/p300, thereby preventing CCND1 transcription(Fig. 2). Thus, the suppression of cyclin D1 as regulated by the ncRNACCND1-TLS complex may participate in G1 arrest in response to DNA damage.
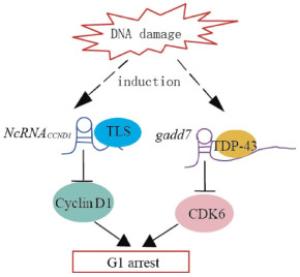
Figure 2. Model demonstrating the proposed mechanisms of lncRNAs-mediated regulation of cyclin D1 and CDK6 induced by DNA damage. DNA damage induces the transcription of ncRNACCND1 from the promoter region of the cyclin D1 gene. NcRNACCND1 associates with and recruits TLS, an RNA-binding protein, to the cyclin D1 promoter, thereby preventing cyclin D1 gene transcription. DNA damage induces the expression of the lncRNAs, gadd7, which dissociates TDP-43 from the CDK6 mRNA to destabilize it, thereby downregulating CDK6 and inhibiting the G1/S transition.
Gadd7 is an lncRNA involved in regulating CDK6 expression[15] in a posttranscriptional manner. TDP-43(TAR DNA binding protein) binds to the 3′ untranslated region of CDK6 mRNA to stabilize it. Gadd7 is transcriptionally induced via the DNA damage mediated by UV radiation and cisplatin[15], binds to TDP-43, and dissociates from CDK6 mRNA[16]. The CDK6 mRNA is then degraded, resulting in the inhibition of the G1/S transition(Fig. 2). Therefore, gadd7 negatively controls CDK6 expression, which functions as a translation regulator.
-
The p21 gene locus expresses several cis- and trans-acting lncRNAs in normal and stressed cells. lncRNA lincRNA-p21 and PANDA are respectively transcribed from 15 kb or 5 kb upstream to the p21 TSS(transcription start site), induced by p53 upon DNA damage, and involved in the regulation in trans of multiple genes downstream of p53[12, 17](Fig. 1). However, the transcriptional regulation of p21 itself is independent of lincRNA-p21 and PANDA[12, 17].
LncRNAs PANDA is a bidirectional transcript from the CDKN1A promoter. PANDA is a p53 effector in response to DNA damage to suppress apoptosis, while CDKN1A induces cell cycle arrest. Suppression of PANDA leads to doxo-induced apoptosis; it simultaneously upregulates genes involved in apoptosis, including the apoptotic activators APAF1(apoptotic protease activating factor 1), BIK, FAS, and LRDD (leucine-rich repeats and death domain containing)[12]. PANDA RNA mechanistically interacts with the transcription factor NF-YA(nuclear transcription factor Y subunit alpha). Together, they are able to suppress the expression of pro-apoptotic genes that carry putative NF-YA binding sites in their promoter regions. The results suggest that abnormal overexpression of PANDA may suppress apoptosis induced by DNA damage that accumulates and pushes the genome toward carcinogenesis. p53 directly binds to its binding element in the lincRNA-p21 promoter. LincRNA-p21 binds to hnRNP-K(heterogeneous nuclear ribonucleoprotein K) and recruits it to the target genes, but the mechanism of target gene regulation is unknown. p53 function is partially mediated by gene regulation via lincRNA-p21-hnRNP-K(Fig. 1).
-
Apart from investigating canonical protein components, recent studies have revealed essential functions of ncRNAs in DDR[18]. Considering that DNA repair factors, such as 53BP1, KU80(a protein that, in humans, is encoded by the XRCC5 gene), and BRCA1, associate with RNA[19-21]; that certain RNA-binding proteins, such as RBMX(RNA binding motif protein, X-linked) and hRNPU(heterogeneous nuclear ribonu-cleoprotein U(scaffold attachment factor A), are re-cruited to DSB sites[4, 22]; and that telomeric repeat-containing RNA associates with DNA repair proteins[23-24], researchers are convinced that DNA damage-induced lncRNAs tend to play a role in DDR.
Wan et al.[25] identified a novel lncRNAs named lncRNAs-JADE. The uniqueness of lncRNAs-JADE lies in the dependence of its induction on ATM but not on p53. Consistent with this finding, no p53-responsive elements are found in the putative promoter region of lncRNAs-JADE. Inhibiting ATM or NF-κB markedly suppresses the DNA damage induction of lncRNAs-JADE and the positive control IRF-1(interferon regulatory factor 1, an NF-κB target). This finding suggests that the transactivation of lncRNA-JADE is regulated by the ATM-NF-κB signaling in the DDR, whereas p53 is dispensable in the induction of lncRNAs-JADE in the DDR.
Furthermore, Jade1 is an essential component of the HBO1(human acetylase binding to ORC1) complex responsible for histone H4 acetylation. Similar to lncRNAs-JADE, the Jade1 protein is further induced after DNA damage. Thus, lncRNAs-JADE is an important link that connects the DNA damage signaling to the Jade1-mediated H4 acetylation[25]. Inhibition of lncRNAs-JADE may enhance the DNA damage-induced apoptosis and may sensitize cells to DNA-damaging agents. Prensner et al.[26] reported the characterization of PCAT-1(prostate cancer associated transcripts) as a prostate cancer lncRNAs implicated in the regulation of DSBs repair. PCAT-1 represses the BRCA2 tumor suppressor gene, thereby leading to downstream impairment of homologous recombination.
-
At present, studies on the functions of lncRNAs mainly focus on tumor, nerve, and development. Thus far, our understanding of the interaction between lncRNAs and DDR, a complex biological problem, remains incomplete. Some key issues are unresolved, such as the origin of lncRNAs, their active mechanism, and the cause of their appearance at the DSBs site. The most important problem is how lncRNAs function at the DNA damage site. Available scarce data suggest that ncRNAs recruit DNA repair proteins to the DSBs site or maintain the DNA repair foci[18]. Intensive study regarding the participation of lncRNAs molecules in the regulatory mechanism of DSBs repair will help us further clarify the mechanism of DNA damage repair. In-depth research can simultaneously provide new guidelines in developing new drugs for radiation protection and tumor chemotherapy or radiosensitivity via the DNA damage repair pathway.
Long Non-coding RNAs in Response to Genotoxic Stress
Long Non-coding RNAs in Response to Genotoxic Stress
-
Abstract: Long non-coding RNAs(lncRNAs) are increasingly involved in diverse biological processes. Upon DNA damage, the DNA damage response(DDR) elicits a complex signaling cascade, which includes the induction of lncRNAs. LncRNA-mediated DDR is involved in non-canonical and canonical manners. DNA-damage induced lncRNAs contribute to the regulation of cell cycle, apoptosis, and DNA repair, thereby playing a key role in maintaining genome stability. This review summarizes the emerging role of lncRNAs in DNA damage and repair.
-
Key words:
- Gene expression /
- Long non-coding RNA /
- DNA damage response /
- Regulatory mechanisms
-
Figure 1. Model demonstrating the proposed mechanisms of lncRNA-mediated regulation of the p53 pathway, which controls cell cycle arrest, repair, and apoptosis in response to DNA damage. In response to DNA damage, p53 is stabilized and activated via phosphorylation mediated by the ATM/ATR pathway. p53 directly binds the target genes and regulates their expression to control cell cycle arrest, repair, and apoptosis. p21 and lincRNA-p21, which are transcribed near the p21 gene, are p53-target genes. LincRNA-p21 controls the expression of some p53-target genes. p53 function is partially mediated by gene regulation via lincRNA-p21.
Figure 2. Model demonstrating the proposed mechanisms of lncRNAs-mediated regulation of cyclin D1 and CDK6 induced by DNA damage. DNA damage induces the transcription of ncRNACCND1 from the promoter region of the cyclin D1 gene. NcRNACCND1 associates with and recruits TLS, an RNA-binding protein, to the cyclin D1 promoter, thereby preventing cyclin D1 gene transcription. DNA damage induces the expression of the lncRNAs, gadd7, which dissociates TDP-43 from the CDK6 mRNA to destabilize it, thereby downregulating CDK6 and inhibiting the G1/S transition.
-
[1] Ciccia A, Elledge SJ. The DNA damage response:making it safe to play with knives[J]. Mol Cell, 2010, 40(2):179-204. DOI:10.1016/j.molcel.2010.09.019. [2] San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination[J]. Annu Rev Biochem, 2008, 77:229-257. DOI:10.1146/annurev.biochem.77.061306.125255. [3] Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway[J]. Annu Rev Biochem, 2010, 79:181-211. DOI:10.1146/annurev.biochem.052308.093131. [4] Polo SE, Blackford AN, Chapman JR, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair[J]. Mol Cell, 2012, 45(4):505-516. DOI:10.1016/j.molcel.2011.12.035. [5] Landau DA, Slack FJ. MicroRNAs in mutagenesis, genomic instability, and DNA repair[J]. Semin Oncol, 2011, 38(6):743-751. DOI:10.1053/j.seminoncol.2011.08.003. [6] Wan GH, Mathur R, Hu XX, et al. miRNA response to DNA damage[J]. Trends Biochem Sci, 2011, 36(9):478-484. DOI:10.1016/j.tibs.2011.06.002. [7] Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription[J]. Nature, 2008, 454(720):126-130. DOI:10.1038/nature06992. [8] Sharma V, Misteli T. Non-coding RNAs in DNA damage and repair[J]. FEBS Lett, 2013, 587(13):1832-1839. DOI:10.1016/j.febslet.2013.05.006. [9] Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage[J]. Science, 2007, 316(5828):1160-1166. DOI:10.1126/science.1140321. [10] Toledo F, Wahl GM. Regulating the p53 pathway:in vitro hypotheses, in vivo veritas[J]. Nat Rev Cancer, 2006, 6(12):909-923. DOI:10.1038/nrc2012. [11] Gibb EA, Vucic EA, Enfield KS, et al. Human cancer long non-coding RNA transcriptomes[J/OL]. PLoS One, 2011, 6(10): e25915[2015-11-24]. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0025915. DOI: 10.1371/journal.pone.0025915. [12] Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters[J]. Nat Genet, 2011, 43(7):621-629. DOI:10.1038/ng.848. [13] Kitagawa M, Kitagawa K, Kotake Y, et al. Cell cycle regulation by long non-coding RNAs[J]. Cell Mol Life Sci, 2013, 70(24):4785-4794. DOI:10.1007/s00018-013-1423-0. [14] Kurokawa R. Promoter-associated long noncoding RNAs repress transcription through a RNA binding protein TLS[J]. Adv Exp Med Biol, 2011, 722:196-208. DOI:10.1007/978-1-4614-0332-6_12. [15] Liu X, Li D, Zhang W, et al. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay[J]. EMBO J, 2012, 31(23):4415-4427. DOI:10.1038/emboj.2012.292. [16] Rossi MN, Antonangeli F. LncRNAs: new players in apoptosis control[J/OL]. Int J Cell Biol, 2014, 2014: 473857[2015-11-24]. http://www.hindawi.com/journals/ijcb/2014/473857. DOI: 10.1155/2014/473857. [17] Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response[J]. Cell, 2010, 142(3):409-419. DOI:10.1016/j.cell.2010.06.040. [18] Zhang C, Peng G. Non-coding RNAs:an emerging player in DNA damage response[J]. Mutat Res Rev Mutat Res, 2015, 763:202-211. DOI:10.1016/j.mrrev.2014.11.003. [19] Mok MT, Henderson BR. Three-dimensional imaging reveals the spatial separation of γH2AX-MDC1-53BP1 and RNF8-RNF168-BRCA1-A complexes at ionizing radiation-induced foci[J]. Radiother Oncol, 2012, 103(3):415-420. DOI:10.1016/j.radonc.2012.04.009. [20] Pfingsten JS, Goodrich KJ, Taabazuing C, et al. Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model[J]. Cell, 2012, 148(5):922-932. DOI:10.1016/j.cell.2012.01.033. [21] Lee YH, Kuo CY, Stark JM, et al. HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response[J]. Nucleic Acids Res, 2013, 41(11):5784-5798. DOI:10.1093/nar/gkt231. [22] Adamson B, Smogorzewska A, Sigoillot FD, et al. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response[J]. Nat Cell Biol, 2012, 14(3):318-328. DOI:10.1038/ncb2426. [23] Nguyen D, Grenier St-Sauveur V, Bergeron D, et al. A Polyadenylation-Dependent 3' end maturation pathway is required for the synthesis of the human telomerase RNA[J]. Cell Rep, 2015, 13(10):2244-2257. DOI:10.1016/j.celrep.2015.11.003. [24] Schoeftner S, Blasco MA. Chromatin regulation and non-coding RNAs at mammalian telomeres[J]. Semin Cell Dev Biol, 2010, 21(2):186-193. DOI:10.1016/j.semcdb.2009.09.015. [25] Wan G, Hu X, Liu Y, et al. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation[J]. EMBO J, 2013, 32(21):2833-2847. DOI:10.1038/emboj.2013.221. [26] Prensner JR, Chen W, Iyer MK, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer[J]. Cancer Res, 2014, 74(6):1651-1660. DOI:10.1158/0008-5472.CAN-13-3159. -




 下载:
下载: